diff --git a/nmpc/newton/README.md b/nmpc/newton/README.md
new file mode 100644
index 0000000..a46901c
--- /dev/null
+++ b/nmpc/newton/README.md
@@ -0,0 +1,79 @@
+# CGMRES method of Nonlinear Model Predictive Control
+This program is about Continuous gmres method for NMPC
+Although usually we have to calculate the partial differential of optimal matrix, it could be really complicated.
+By using CGMRES, we can pass the calculating step and get the optimal input quickly.
+
+# Problem Formulation
+
+- **example**
+
+- model
+
+x_2-x_1+u&space;\\&space;\end{bmatrix},&space;|u|&space;\leq&space;0.5) +
+- evaluation function
+
+
+
+- evaluation function
+
++x_2^2(t+T))+\int_{t}^{t+T}\frac{1}{2}(x_1^2+x_2^2+u^2)-0.01vd\tau) +
+
+- **two wheeled model**
+
+- model
+
+
+
+
+- **two wheeled model**
+
+- model
+
+&space;&&space;0&space;\\&space;\sin(\theta)&space;&&space;0&space;\\&space;0&space;&&space;1&space;\\&space;\end{bmatrix}&space;\begin{bmatrix}&space;u_v&space;\\&space;u_\omega&space;\\&space;\end{bmatrix}&space;=&space;\boldsymbol{B}\boldsymbol{U}) +
+- evaluation function
+
+
+
+- evaluation function
+
+^2&space;+&space;\int_{t_0}^{t_0&space;+&space;T}&space;\boldsymbol{U}(t)^2&space;-&space;0.01&space;dummy_{u_v}&space;-&space;dummy_{u_\omega}&space;dt) +
+
+if you want to see more detail about this methods, you should go https://qiita.com/MENDY/items/4108190a579395053924.
+However, it is written in Japanese
+
+# Expected Results
+
+- example
+
+
+
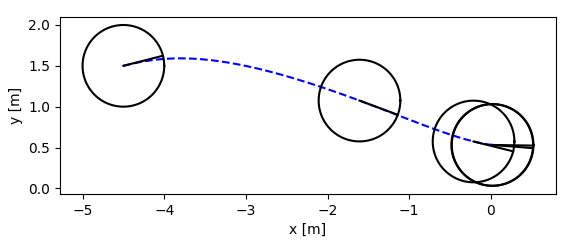
+- two wheeled model
+
+- trajectory
+
+
+
+- time history
+
+
+
+
+# Usage
+
+- for example
+
+```
+$ python main_example.py
+```
+
+- for two wheeled
+
+```
+$ python main_two_wheeled.py
+```
+
+# Requirement
+
+- python3.5 or more
+- numpy
+- matplotlib
+
+# Reference
+I`m sorry that main references are written in Japanese
+
+- main (commentary article) (Japanse) https://qiita.com/MENDY/items/4108190a579395053924
+
+- Ohtsuka, T., & Fujii, H. A. (1997). Real-time Optimization Algorithm for Nonlinear Receding-horizon Control. Automatica, 33(6), 1147–1154. https://doi.org/10.1016/S0005-1098(97)00005-8
+
+- 非線形最適制御入門(コロナ社)
+
+- 実時間最適化による制御の実応用(コロナ社)
\ No newline at end of file
diff --git a/nmpc/newton/main_example.py b/nmpc/newton/main_example.py
new file mode 100644
index 0000000..06d8c17
--- /dev/null
+++ b/nmpc/newton/main_example.py
@@ -0,0 +1,647 @@
+import numpy as np
+import matplotlib.pyplot as plt
+import math
+
+class SampleSystem():
+ """SampleSystem, this is the simulator
+ Attributes
+ -----------
+ x_1 : float
+ system state 1
+ x_2 : float
+ system state 2
+ history_x_1 : list
+ time history of system state 1 (x_1)
+ history_x_2 : list
+ time history of system state 2 (x_2)
+ """
+ def __init__(self, init_x_1=0., init_x_2=0.):
+ """
+ Palameters
+ -----------
+ init_x_1 : float, optional
+ initial value of x_1, default is 0.
+ init_x_2 : float, optional
+ initial value of x_2, default is 0.
+ """
+ self.x_1 = init_x_1
+ self.x_2 = init_x_2
+ self.history_x_1 = [init_x_1]
+ self.history_x_2 = [init_x_2]
+
+ def update_state(self, u, dt=0.01):
+ """
+ Palameters
+ ------------
+ u : float
+ input of system in some cases this means the reference
+ dt : float in seconds, optional
+ sampling time of simulation, default is 0.01 [s]
+ """
+ # for theta 1, theta 1 dot, theta 2, theta 2 dot
+ k0 = [0.0 for _ in range(2)]
+ k1 = [0.0 for _ in range(2)]
+ k2 = [0.0 for _ in range(2)]
+ k3 = [0.0 for _ in range(2)]
+
+ functions = [self._func_x_1, self._func_x_2]
+
+ # solve Runge-Kutta
+ for i, func in enumerate(functions):
+ k0[i] = dt * func(self.x_1, self.x_2, u)
+
+ for i, func in enumerate(functions):
+ k1[i] = dt * func(self.x_1 + k0[0]/2., self.x_2 + k0[1]/2., u)
+
+ for i, func in enumerate(functions):
+ k2[i] = dt * func(self.x_1 + k1[0]/2., self.x_2 + k1[1]/2., u)
+
+ for i, func in enumerate(functions):
+ k3[i] = dt * func(self.x_1 + k2[0], self.x_2 + k2[1], u)
+
+ self.x_1 += (k0[0] + 2. * k1[0] + 2. * k2[0] + k3[0]) / 6.
+ self.x_2 += (k0[1] + 2. * k1[1] + 2. * k2[1] + k3[1]) / 6.
+
+ # save
+ self.history_x_1.append(self.x_1)
+ self.history_x_2.append(self.x_2)
+
+ def _func_x_1(self, y_1, y_2, u):
+ """
+ Parameters
+ ------------
+ y_1 : float
+ y_2 : float
+ u : float
+ system input
+ """
+ y_dot = y_2
+ return y_dot
+
+ def _func_x_2(self, y_1, y_2, u):
+ """
+ Parameters
+ ------------
+ y_1 : float
+ y_2 : float
+ u : float
+ system input
+ """
+ y_dot = (1. - y_1**2 - y_2**2) * y_2 - y_1 + u
+ return y_dot
+
+
+class NMPCSimulatorSystem():
+ """SimulatorSystem for nmpc, this is the simulator of nmpc
+ the reason why I seperate the real simulator and nmpc's simulator is sometimes the modeling error, disturbance can include in real simulator
+ Attributes
+ -----------
+
+ """
+ def __init__(self):
+ """
+ Parameters
+ -----------
+ None
+ """
+ pass
+
+ def calc_predict_and_adjoint_state(self, x_1, x_2, us, N, dt):
+ """main
+ Parameters
+ ------------
+ x_1 : float
+ current state
+ x_2 : float
+ current state
+ us : list of float
+ estimated optimal input Us for N steps
+ N : int
+ predict step
+ dt : float
+ sampling time
+
+ Returns
+ --------
+ x_1s : list of float
+ predicted x_1s for N steps
+ x_2s : list of float
+ predicted x_2s for N steps
+ lam_1s : list of float
+ adjoint state of x_1s, lam_1s for N steps
+ lam_2s : list of float
+ adjoint state of x_2s, lam_2s for N steps
+ """
+
+ x_1s, x_2s = self._calc_predict_states(x_1, x_2, us, N, dt) # by usin state equation
+ lam_1s, lam_2s = self._calc_adjoint_states(x_1s, x_2s, us, N, dt) # by using adjoint equation
+
+ return x_1s, x_2s, lam_1s, lam_2s
+
+ def _calc_predict_states(self, x_1, x_2, us, N, dt):
+ """
+ Parameters
+ ------------
+ x_1 : float
+ current state
+ x_2 : float
+ current state
+ us : list of float
+ estimated optimal input Us for N steps
+ N : int
+ predict step
+ dt : float
+ sampling time
+
+ Returns
+ --------
+ x_1s : list of float
+ predicted x_1s for N steps
+ x_2s : list of float
+ predicted x_2s for N steps
+ """
+ # initial state
+ x_1s = [x_1]
+ x_2s = [x_2]
+
+ for i in range(N):
+ temp_x_1, temp_x_2 = self._predict_state_with_oylar(x_1s[i], x_2s[i], us[i], dt)
+ x_1s.append(temp_x_1)
+ x_2s.append(temp_x_2)
+
+ return x_1s, x_2s
+
+ def _calc_adjoint_states(self, x_1s, x_2s, us, N, dt):
+ """
+ Parameters
+ ------------
+ x_1s : list of float
+ predicted x_1s for N steps
+ x_2s : list of float
+ predicted x_2s for N steps
+ us : list of float
+ estimated optimal input Us for N steps
+ N : int
+ predict step
+ dt : float
+ sampling time
+
+ Returns
+ --------
+ lam_1s : list of float
+ adjoint state of x_1s, lam_1s for N steps
+ lam_2s : list of float
+ adjoint state of x_2s, lam_2s for N steps
+ """
+ # final state
+ # final_state_func
+ lam_1s = [x_1s[-1]]
+ lam_2s = [x_2s[-1]]
+
+ for i in range(N-1, 0, -1):
+ temp_lam_1, temp_lam_2 = self._adjoint_state_with_oylar(x_1s[i], x_2s[i], lam_1s[0] ,lam_2s[0], us[i], dt)
+ lam_1s.insert(0, temp_lam_1)
+ lam_2s.insert(0, temp_lam_2)
+
+ return lam_1s, lam_2s
+
+ def final_state_func(self):
+ """this func usually need
+ """
+ pass
+
+ def _predict_state_with_oylar(self, x_1, x_2, u, dt):
+ """in this case this function is the same as simulator
+ Parameters
+ ------------
+ x_1 : float
+ system state
+ x_2 : float
+ system state
+ u : float
+ system input
+ dt : float in seconds
+ sampling time
+ Returns
+ --------
+ next_x_1 : float
+ next state, x_1 calculated by using state equation
+ next_x_2 : float
+ next state, x_2 calculated by using state equation
+ """
+ k0 = [0. for _ in range(2)]
+
+ functions = [self.func_x_1, self.func_x_2]
+
+ for i, func in enumerate(functions):
+ k0[i] = dt * func(x_1, x_2, u)
+
+ next_x_1 = x_1 + k0[0]
+ next_x_2 = x_2 + k0[1]
+
+ return next_x_1, next_x_2
+
+ def func_x_1(self, y_1, y_2, u):
+ """calculating \dot{x_1}
+ Parameters
+ ------------
+ y_1 : float
+ means x_1
+ y_2 : float
+ means x_2
+ u : float
+ means system input
+ Returns
+ ---------
+ y_dot : float
+ means \dot{x_1}
+ """
+ y_dot = y_2
+ return y_dot
+
+ def func_x_2(self, y_1, y_2, u):
+ """calculating \dot{x_2}
+ Parameters
+ ------------
+ y_1 : float
+ means x_1
+ y_2 : float
+ means x_2
+ u : float
+ means system input
+ Returns
+ ---------
+ y_dot : float
+ means \dot{x_2}
+ """
+ y_dot = (1. - y_1**2 - y_2**2) * y_2 - y_1 + u
+ return y_dot
+
+ def _adjoint_state_with_oylar(self, x_1, x_2, lam_1, lam_2, u, dt):
+ """
+ Parameters
+ ------------
+ x_1 : float
+ system state
+ x_2 : float
+ system state
+ lam_1 : float
+ adjoint state
+ lam_2 : float
+ adjoint state
+ u : float
+ system input
+ dt : float in seconds
+ sampling time
+ Returns
+ --------
+ pre_lam_1 : float
+ pre, 1 step before lam_1 calculated by using adjoint equation
+ pre_lam_2 : float
+ pre, 1 step before lam_2 calculated by using adjoint equation
+ """
+ k0 = [0. for _ in range(2)]
+
+ functions = [self._func_lam_1, self._func_lam_2]
+
+ for i, func in enumerate(functions):
+ k0[i] = dt * func(x_1, x_2, lam_1, lam_2, u)
+
+ pre_lam_1 = lam_1 + k0[0]
+ pre_lam_2 = lam_2 + k0[1]
+
+ return pre_lam_1, pre_lam_2
+
+ def _func_lam_1(self, y_1, y_2, y_3, y_4, u):
+ """calculating -\dot{lam_1}
+ Parameters
+ ------------
+ y_1 : float
+ means x_1
+ y_2 : float
+ means x_2
+ y_3 : float
+ means lam_1
+ y_4 : float
+ means lam_2
+ u : float
+ means system input
+ Returns
+ ---------
+ y_dot : float
+ means -\dot{lam_1}
+ """
+ y_dot = y_1 - (2. * y_1 * y_2 + 1.) * y_4
+ return y_dot
+
+ def _func_lam_2(self, y_1, y_2, y_3, y_4, u):
+ """calculating -\dot{lam_2}
+ Parameters
+ ------------
+ y_1 : float
+ means x_1
+ y_2 : float
+ means x_2
+ y_3 : float
+ means lam_1
+ y_4 : float
+ means lam_2
+ u : float
+ means system input
+ Returns
+ ---------
+ y_dot : float
+ means -\dot{lam_2}
+ """
+ y_dot = y_2 + y_3 + (-3. * (y_2**2) - y_1**2 + 1. ) * y_4
+ return y_dot
+
+
+def calc_numerical_gradient(f, x, shape):
+ """
+ Parameters
+ ------------
+ f : function
+ forward function of NN
+ x : numpy.ndarray
+ input
+ shape : tuple
+ jacobian shape
+
+ Returns
+ ---------
+ grad : numpy.ndarray, shape is the same as shape
+ results of numercial gradient of the input
+ References
+ -----------
+ - oreilly japan 0 から作るdeeplearning
+ https://github.com/oreilly-japan/deep-learning-from-scratch/blob/master/common/gradient.py
+ """
+ # check condition
+ if not callable(f):
+ raise TypeError("f should be callable")
+
+ if not (isinstance(x, list) or isinstance(x, np.ndarray)):
+ raise TypeError("x should be array-like")
+
+ h = 1e-3 # 0.01
+ grad = np.zeros(shape)
+
+ for idx in range(x.size):
+ # save
+ tmp_val = x[idx]
+
+ x[idx] = float(tmp_val) + h
+ fxh1 = f(x) # f(x+h)
+
+ x[idx] = float(tmp_val) - h
+ fxh2 = f(x) # f(x-h)
+
+ grad[:, idx] = (fxh1 - fxh2) / (2*h)
+
+ x[idx] = tmp_val
+
+ return np.array(grad)
+
+class NMPCControllerWithNewton():
+ """
+ Attributes
+ ------------
+ zeta : float
+ gain of optimal answer stability
+ tf : float
+ predict time
+ alpha : float
+ gain of predict time
+ N : int
+ predicte step, discritize value
+ threshold : float
+ cgmres's threshold value
+ input_num : int
+ system input length, this should include dummy u and constraint variables
+ max_iteration : int
+ decide by the solved matrix size
+ simulator : NMPCSimulatorSystem class
+ us : list of float
+ estimated optimal system input
+ dummy_us : list of float
+ estimated dummy input
+ raws : list of float
+ estimated constraint variable
+ history_u : list of float
+ time history of actual system input
+ history_dummy_u : list of float
+ time history of actual dummy u
+ history_raw : list of float
+ time history of actual raw
+ history_f : list of float
+ time history of error of optimal
+ """
+ def __init__(self):
+ """
+ Parameters
+ -----------
+ None
+ """
+ # parameters
+ self.tf = 1. # 最終時間
+ self.N = 10 # 分割数
+ self.threshold = 0.0001 # break値
+
+ self.NUM_INPUT = 3 # dummy, 制約条件に対するrawにも合わせた入力の数
+ self.Jacobian_size = self.NUM_INPUT * self.N
+
+ # newton parameters
+ self.MAX_ITERATION = 100
+
+ # simulator
+ self.simulator = NMPCSimulatorSystem()
+
+ # initial
+ self.us = np.zeros(self.N)
+ self.dummy_us = np.ones(self.N) * 0.25
+ self.raws = np.ones(self.N) * 0.01
+
+ # for fig
+ self.history_u = []
+ self.history_dummy_u = []
+ self.history_raw = []
+ self.history_f = []
+
+ def calc_input(self, x_1, x_2, time):
+ """
+ Parameters
+ ------------
+ x_1 : float
+ current state
+ x_2 : float
+ current state
+ time : float in seconds
+ now time
+ Returns
+ --------
+ us : list of float
+ estimated optimal system input
+ """
+ # calculating sampling time
+ dt = 0.01
+
+ # concat all us, shape (NUM_INPUT, N)
+ all_us = np.stack((self.us, self.dummy_us, self.raws))
+ all_us = all_us.T.flatten()
+
+ # Newton method
+ for i in range(self.MAX_ITERATION):
+ # check
+ # print("all_us = {}".format(all_us))
+ # print("newton iteration in {}".format(i))
+ # input()
+
+ # calc all state
+ x_1s, x_2s, lam_1s, lam_2s = self.simulator.calc_predict_and_adjoint_state(x_1, x_2, self.us, self.N, dt)
+
+ # F
+ F_hat = self._calc_f(x_1s, x_2s, lam_1s, lam_2s, all_us, self.N, dt)
+
+ # judge
+ # print("F_hat = {}".format(F_hat))
+ # print(np.linalg.norm(F_hat))
+ if np.linalg.norm(F_hat) < self.threshold:
+ # print("break!!")
+ break
+
+ grad_f = lambda all_us : self._calc_f(x_1s, x_2s, lam_1s, lam_2s, all_us, self.N, dt)
+ grads = calc_numerical_gradient(grad_f, all_us, (self.Jacobian_size, self.Jacobian_size))
+
+ # make jacobian and calc inverse of it
+ # all us
+ all_us = all_us - np.dot(np.linalg.inv(grads), F_hat)
+
+ # update
+ self.us = all_us[::self.NUM_INPUT]
+ self.dummy_us = all_us[1::self.NUM_INPUT]
+ self.raws = all_us[2::self.NUM_INPUT]
+
+ # final insert
+ self.us = all_us[::self.NUM_INPUT]
+ self.dummy_us = all_us[1::self.NUM_INPUT]
+ self.raws = all_us[2::self.NUM_INPUT]
+
+ x_1s, x_2s, lam_1s, lam_2s = self.simulator.calc_predict_and_adjoint_state(x_1, x_2, self.us, self.N, dt)
+
+ F = self._calc_f(x_1s, x_2s, lam_1s, lam_2s, all_us, self.N, dt)
+
+ # print("check val of F = {0}".format(np.linalg.norm(F)))
+ # input()
+
+ # for save
+ self.history_f.append(np.linalg.norm(F))
+ self.history_u.append(self.us[0])
+ self.history_dummy_u.append(self.dummy_us[0])
+ self.history_raw.append(self.raws[0])
+
+ return self.us
+
+ def _calc_f(self, x_1s, x_2s, lam_1s, lam_2s, all_us, N, dt):
+ """
+ Parameters
+ ------------
+ x_1s : list of float
+ predicted x_1s for N steps
+ x_2s : list of float
+ predicted x_2s for N steps
+ lam_1s : list of float
+ adjoint state of x_1s, lam_1s for N steps
+ lam_2s : list of float
+ adjoint state of x_2s, lam_2s for N steps
+ us : list of float
+ estimated optimal system input
+ dummy_us : list of float
+ estimated dummy input
+ raws : list of float
+ estimated constraint variable
+ N : int
+ predict time step
+ dt : float
+ sampling time of system
+ """
+ F = []
+
+ us = all_us[::self.NUM_INPUT]
+ dummy_us = all_us[1::self.NUM_INPUT]
+ raws = all_us[2::self.NUM_INPUT]
+
+ for i in range(N):
+ F.append(0.5 * us[i] + lam_2s[i] + 2. * raws[i] * us[i])
+ F.append(-0.01 + 2. * raws[i] * dummy_us[i])
+ F.append(us[i]**2 + dummy_us[i]**2 - 0.5**2)
+
+ return np.array(F)
+
+def main():
+ # simulation time
+ dt = 0.01
+ iteration_time = 20.
+ iteration_num = int(iteration_time/dt)
+
+ # plant
+ plant_system = SampleSystem(init_x_1=2., init_x_2=0.)
+
+ # controller
+ controller = NMPCControllerWithNewton()
+
+ # for i in range(iteration_num)
+ for i in range(1, iteration_num):
+ print("iteration = {}".format(i))
+ time = float(i) * dt
+ x_1 = plant_system.x_1
+ x_2 = plant_system.x_2
+ # make input
+ us = controller.calc_input(x_1, x_2, time)
+ # update state
+ plant_system.update_state(us[0])
+
+ # figure
+ fig = plt.figure()
+
+ x_1_fig = fig.add_subplot(321)
+ x_2_fig = fig.add_subplot(322)
+ u_fig = fig.add_subplot(323)
+ dummy_fig = fig.add_subplot(324)
+ raw_fig = fig.add_subplot(325)
+ f_fig = fig.add_subplot(326)
+
+ x_1_fig.plot(np.arange(iteration_num)*dt, plant_system.history_x_1)
+ x_1_fig.set_xlabel("time [s]")
+ x_1_fig.set_ylabel("x_1")
+
+ x_2_fig.plot(np.arange(iteration_num)*dt, plant_system.history_x_2)
+ x_2_fig.set_xlabel("time [s]")
+ x_2_fig.set_ylabel("x_2")
+
+ u_fig.plot(np.arange(iteration_num - 1)*dt, controller.history_u)
+ u_fig.set_xlabel("time [s]")
+ u_fig.set_ylabel("u")
+
+ dummy_fig.plot(np.arange(iteration_num - 1)*dt, controller.history_dummy_u)
+ dummy_fig.set_xlabel("time [s]")
+ dummy_fig.set_ylabel("dummy u")
+
+ raw_fig.plot(np.arange(iteration_num - 1)*dt, controller.history_raw)
+ raw_fig.set_xlabel("time [s]")
+ raw_fig.set_ylabel("raw")
+
+ f_fig.plot(np.arange(iteration_num - 1)*dt, controller.history_f)
+ f_fig.set_xlabel("time [s]")
+ f_fig.set_ylabel("optimal error")
+
+ fig.tight_layout()
+
+ plt.show()
+
+
+if __name__ == "__main__":
+ main()
+
+
+
+
+
+if you want to see more detail about this methods, you should go https://qiita.com/MENDY/items/4108190a579395053924.
+However, it is written in Japanese
+
+# Expected Results
+
+- example
+
+
+
+- two wheeled model
+
+- trajectory
+
+
+
+- time history
+
+
+
+
+# Usage
+
+- for example
+
+```
+$ python main_example.py
+```
+
+- for two wheeled
+
+```
+$ python main_two_wheeled.py
+```
+
+# Requirement
+
+- python3.5 or more
+- numpy
+- matplotlib
+
+# Reference
+I`m sorry that main references are written in Japanese
+
+- main (commentary article) (Japanse) https://qiita.com/MENDY/items/4108190a579395053924
+
+- Ohtsuka, T., & Fujii, H. A. (1997). Real-time Optimization Algorithm for Nonlinear Receding-horizon Control. Automatica, 33(6), 1147–1154. https://doi.org/10.1016/S0005-1098(97)00005-8
+
+- 非線形最適制御入門(コロナ社)
+
+- 実時間最適化による制御の実応用(コロナ社)
\ No newline at end of file
diff --git a/nmpc/newton/main_example.py b/nmpc/newton/main_example.py
new file mode 100644
index 0000000..06d8c17
--- /dev/null
+++ b/nmpc/newton/main_example.py
@@ -0,0 +1,647 @@
+import numpy as np
+import matplotlib.pyplot as plt
+import math
+
+class SampleSystem():
+ """SampleSystem, this is the simulator
+ Attributes
+ -----------
+ x_1 : float
+ system state 1
+ x_2 : float
+ system state 2
+ history_x_1 : list
+ time history of system state 1 (x_1)
+ history_x_2 : list
+ time history of system state 2 (x_2)
+ """
+ def __init__(self, init_x_1=0., init_x_2=0.):
+ """
+ Palameters
+ -----------
+ init_x_1 : float, optional
+ initial value of x_1, default is 0.
+ init_x_2 : float, optional
+ initial value of x_2, default is 0.
+ """
+ self.x_1 = init_x_1
+ self.x_2 = init_x_2
+ self.history_x_1 = [init_x_1]
+ self.history_x_2 = [init_x_2]
+
+ def update_state(self, u, dt=0.01):
+ """
+ Palameters
+ ------------
+ u : float
+ input of system in some cases this means the reference
+ dt : float in seconds, optional
+ sampling time of simulation, default is 0.01 [s]
+ """
+ # for theta 1, theta 1 dot, theta 2, theta 2 dot
+ k0 = [0.0 for _ in range(2)]
+ k1 = [0.0 for _ in range(2)]
+ k2 = [0.0 for _ in range(2)]
+ k3 = [0.0 for _ in range(2)]
+
+ functions = [self._func_x_1, self._func_x_2]
+
+ # solve Runge-Kutta
+ for i, func in enumerate(functions):
+ k0[i] = dt * func(self.x_1, self.x_2, u)
+
+ for i, func in enumerate(functions):
+ k1[i] = dt * func(self.x_1 + k0[0]/2., self.x_2 + k0[1]/2., u)
+
+ for i, func in enumerate(functions):
+ k2[i] = dt * func(self.x_1 + k1[0]/2., self.x_2 + k1[1]/2., u)
+
+ for i, func in enumerate(functions):
+ k3[i] = dt * func(self.x_1 + k2[0], self.x_2 + k2[1], u)
+
+ self.x_1 += (k0[0] + 2. * k1[0] + 2. * k2[0] + k3[0]) / 6.
+ self.x_2 += (k0[1] + 2. * k1[1] + 2. * k2[1] + k3[1]) / 6.
+
+ # save
+ self.history_x_1.append(self.x_1)
+ self.history_x_2.append(self.x_2)
+
+ def _func_x_1(self, y_1, y_2, u):
+ """
+ Parameters
+ ------------
+ y_1 : float
+ y_2 : float
+ u : float
+ system input
+ """
+ y_dot = y_2
+ return y_dot
+
+ def _func_x_2(self, y_1, y_2, u):
+ """
+ Parameters
+ ------------
+ y_1 : float
+ y_2 : float
+ u : float
+ system input
+ """
+ y_dot = (1. - y_1**2 - y_2**2) * y_2 - y_1 + u
+ return y_dot
+
+
+class NMPCSimulatorSystem():
+ """SimulatorSystem for nmpc, this is the simulator of nmpc
+ the reason why I seperate the real simulator and nmpc's simulator is sometimes the modeling error, disturbance can include in real simulator
+ Attributes
+ -----------
+
+ """
+ def __init__(self):
+ """
+ Parameters
+ -----------
+ None
+ """
+ pass
+
+ def calc_predict_and_adjoint_state(self, x_1, x_2, us, N, dt):
+ """main
+ Parameters
+ ------------
+ x_1 : float
+ current state
+ x_2 : float
+ current state
+ us : list of float
+ estimated optimal input Us for N steps
+ N : int
+ predict step
+ dt : float
+ sampling time
+
+ Returns
+ --------
+ x_1s : list of float
+ predicted x_1s for N steps
+ x_2s : list of float
+ predicted x_2s for N steps
+ lam_1s : list of float
+ adjoint state of x_1s, lam_1s for N steps
+ lam_2s : list of float
+ adjoint state of x_2s, lam_2s for N steps
+ """
+
+ x_1s, x_2s = self._calc_predict_states(x_1, x_2, us, N, dt) # by usin state equation
+ lam_1s, lam_2s = self._calc_adjoint_states(x_1s, x_2s, us, N, dt) # by using adjoint equation
+
+ return x_1s, x_2s, lam_1s, lam_2s
+
+ def _calc_predict_states(self, x_1, x_2, us, N, dt):
+ """
+ Parameters
+ ------------
+ x_1 : float
+ current state
+ x_2 : float
+ current state
+ us : list of float
+ estimated optimal input Us for N steps
+ N : int
+ predict step
+ dt : float
+ sampling time
+
+ Returns
+ --------
+ x_1s : list of float
+ predicted x_1s for N steps
+ x_2s : list of float
+ predicted x_2s for N steps
+ """
+ # initial state
+ x_1s = [x_1]
+ x_2s = [x_2]
+
+ for i in range(N):
+ temp_x_1, temp_x_2 = self._predict_state_with_oylar(x_1s[i], x_2s[i], us[i], dt)
+ x_1s.append(temp_x_1)
+ x_2s.append(temp_x_2)
+
+ return x_1s, x_2s
+
+ def _calc_adjoint_states(self, x_1s, x_2s, us, N, dt):
+ """
+ Parameters
+ ------------
+ x_1s : list of float
+ predicted x_1s for N steps
+ x_2s : list of float
+ predicted x_2s for N steps
+ us : list of float
+ estimated optimal input Us for N steps
+ N : int
+ predict step
+ dt : float
+ sampling time
+
+ Returns
+ --------
+ lam_1s : list of float
+ adjoint state of x_1s, lam_1s for N steps
+ lam_2s : list of float
+ adjoint state of x_2s, lam_2s for N steps
+ """
+ # final state
+ # final_state_func
+ lam_1s = [x_1s[-1]]
+ lam_2s = [x_2s[-1]]
+
+ for i in range(N-1, 0, -1):
+ temp_lam_1, temp_lam_2 = self._adjoint_state_with_oylar(x_1s[i], x_2s[i], lam_1s[0] ,lam_2s[0], us[i], dt)
+ lam_1s.insert(0, temp_lam_1)
+ lam_2s.insert(0, temp_lam_2)
+
+ return lam_1s, lam_2s
+
+ def final_state_func(self):
+ """this func usually need
+ """
+ pass
+
+ def _predict_state_with_oylar(self, x_1, x_2, u, dt):
+ """in this case this function is the same as simulator
+ Parameters
+ ------------
+ x_1 : float
+ system state
+ x_2 : float
+ system state
+ u : float
+ system input
+ dt : float in seconds
+ sampling time
+ Returns
+ --------
+ next_x_1 : float
+ next state, x_1 calculated by using state equation
+ next_x_2 : float
+ next state, x_2 calculated by using state equation
+ """
+ k0 = [0. for _ in range(2)]
+
+ functions = [self.func_x_1, self.func_x_2]
+
+ for i, func in enumerate(functions):
+ k0[i] = dt * func(x_1, x_2, u)
+
+ next_x_1 = x_1 + k0[0]
+ next_x_2 = x_2 + k0[1]
+
+ return next_x_1, next_x_2
+
+ def func_x_1(self, y_1, y_2, u):
+ """calculating \dot{x_1}
+ Parameters
+ ------------
+ y_1 : float
+ means x_1
+ y_2 : float
+ means x_2
+ u : float
+ means system input
+ Returns
+ ---------
+ y_dot : float
+ means \dot{x_1}
+ """
+ y_dot = y_2
+ return y_dot
+
+ def func_x_2(self, y_1, y_2, u):
+ """calculating \dot{x_2}
+ Parameters
+ ------------
+ y_1 : float
+ means x_1
+ y_2 : float
+ means x_2
+ u : float
+ means system input
+ Returns
+ ---------
+ y_dot : float
+ means \dot{x_2}
+ """
+ y_dot = (1. - y_1**2 - y_2**2) * y_2 - y_1 + u
+ return y_dot
+
+ def _adjoint_state_with_oylar(self, x_1, x_2, lam_1, lam_2, u, dt):
+ """
+ Parameters
+ ------------
+ x_1 : float
+ system state
+ x_2 : float
+ system state
+ lam_1 : float
+ adjoint state
+ lam_2 : float
+ adjoint state
+ u : float
+ system input
+ dt : float in seconds
+ sampling time
+ Returns
+ --------
+ pre_lam_1 : float
+ pre, 1 step before lam_1 calculated by using adjoint equation
+ pre_lam_2 : float
+ pre, 1 step before lam_2 calculated by using adjoint equation
+ """
+ k0 = [0. for _ in range(2)]
+
+ functions = [self._func_lam_1, self._func_lam_2]
+
+ for i, func in enumerate(functions):
+ k0[i] = dt * func(x_1, x_2, lam_1, lam_2, u)
+
+ pre_lam_1 = lam_1 + k0[0]
+ pre_lam_2 = lam_2 + k0[1]
+
+ return pre_lam_1, pre_lam_2
+
+ def _func_lam_1(self, y_1, y_2, y_3, y_4, u):
+ """calculating -\dot{lam_1}
+ Parameters
+ ------------
+ y_1 : float
+ means x_1
+ y_2 : float
+ means x_2
+ y_3 : float
+ means lam_1
+ y_4 : float
+ means lam_2
+ u : float
+ means system input
+ Returns
+ ---------
+ y_dot : float
+ means -\dot{lam_1}
+ """
+ y_dot = y_1 - (2. * y_1 * y_2 + 1.) * y_4
+ return y_dot
+
+ def _func_lam_2(self, y_1, y_2, y_3, y_4, u):
+ """calculating -\dot{lam_2}
+ Parameters
+ ------------
+ y_1 : float
+ means x_1
+ y_2 : float
+ means x_2
+ y_3 : float
+ means lam_1
+ y_4 : float
+ means lam_2
+ u : float
+ means system input
+ Returns
+ ---------
+ y_dot : float
+ means -\dot{lam_2}
+ """
+ y_dot = y_2 + y_3 + (-3. * (y_2**2) - y_1**2 + 1. ) * y_4
+ return y_dot
+
+
+def calc_numerical_gradient(f, x, shape):
+ """
+ Parameters
+ ------------
+ f : function
+ forward function of NN
+ x : numpy.ndarray
+ input
+ shape : tuple
+ jacobian shape
+
+ Returns
+ ---------
+ grad : numpy.ndarray, shape is the same as shape
+ results of numercial gradient of the input
+ References
+ -----------
+ - oreilly japan 0 から作るdeeplearning
+ https://github.com/oreilly-japan/deep-learning-from-scratch/blob/master/common/gradient.py
+ """
+ # check condition
+ if not callable(f):
+ raise TypeError("f should be callable")
+
+ if not (isinstance(x, list) or isinstance(x, np.ndarray)):
+ raise TypeError("x should be array-like")
+
+ h = 1e-3 # 0.01
+ grad = np.zeros(shape)
+
+ for idx in range(x.size):
+ # save
+ tmp_val = x[idx]
+
+ x[idx] = float(tmp_val) + h
+ fxh1 = f(x) # f(x+h)
+
+ x[idx] = float(tmp_val) - h
+ fxh2 = f(x) # f(x-h)
+
+ grad[:, idx] = (fxh1 - fxh2) / (2*h)
+
+ x[idx] = tmp_val
+
+ return np.array(grad)
+
+class NMPCControllerWithNewton():
+ """
+ Attributes
+ ------------
+ zeta : float
+ gain of optimal answer stability
+ tf : float
+ predict time
+ alpha : float
+ gain of predict time
+ N : int
+ predicte step, discritize value
+ threshold : float
+ cgmres's threshold value
+ input_num : int
+ system input length, this should include dummy u and constraint variables
+ max_iteration : int
+ decide by the solved matrix size
+ simulator : NMPCSimulatorSystem class
+ us : list of float
+ estimated optimal system input
+ dummy_us : list of float
+ estimated dummy input
+ raws : list of float
+ estimated constraint variable
+ history_u : list of float
+ time history of actual system input
+ history_dummy_u : list of float
+ time history of actual dummy u
+ history_raw : list of float
+ time history of actual raw
+ history_f : list of float
+ time history of error of optimal
+ """
+ def __init__(self):
+ """
+ Parameters
+ -----------
+ None
+ """
+ # parameters
+ self.tf = 1. # 最終時間
+ self.N = 10 # 分割数
+ self.threshold = 0.0001 # break値
+
+ self.NUM_INPUT = 3 # dummy, 制約条件に対するrawにも合わせた入力の数
+ self.Jacobian_size = self.NUM_INPUT * self.N
+
+ # newton parameters
+ self.MAX_ITERATION = 100
+
+ # simulator
+ self.simulator = NMPCSimulatorSystem()
+
+ # initial
+ self.us = np.zeros(self.N)
+ self.dummy_us = np.ones(self.N) * 0.25
+ self.raws = np.ones(self.N) * 0.01
+
+ # for fig
+ self.history_u = []
+ self.history_dummy_u = []
+ self.history_raw = []
+ self.history_f = []
+
+ def calc_input(self, x_1, x_2, time):
+ """
+ Parameters
+ ------------
+ x_1 : float
+ current state
+ x_2 : float
+ current state
+ time : float in seconds
+ now time
+ Returns
+ --------
+ us : list of float
+ estimated optimal system input
+ """
+ # calculating sampling time
+ dt = 0.01
+
+ # concat all us, shape (NUM_INPUT, N)
+ all_us = np.stack((self.us, self.dummy_us, self.raws))
+ all_us = all_us.T.flatten()
+
+ # Newton method
+ for i in range(self.MAX_ITERATION):
+ # check
+ # print("all_us = {}".format(all_us))
+ # print("newton iteration in {}".format(i))
+ # input()
+
+ # calc all state
+ x_1s, x_2s, lam_1s, lam_2s = self.simulator.calc_predict_and_adjoint_state(x_1, x_2, self.us, self.N, dt)
+
+ # F
+ F_hat = self._calc_f(x_1s, x_2s, lam_1s, lam_2s, all_us, self.N, dt)
+
+ # judge
+ # print("F_hat = {}".format(F_hat))
+ # print(np.linalg.norm(F_hat))
+ if np.linalg.norm(F_hat) < self.threshold:
+ # print("break!!")
+ break
+
+ grad_f = lambda all_us : self._calc_f(x_1s, x_2s, lam_1s, lam_2s, all_us, self.N, dt)
+ grads = calc_numerical_gradient(grad_f, all_us, (self.Jacobian_size, self.Jacobian_size))
+
+ # make jacobian and calc inverse of it
+ # all us
+ all_us = all_us - np.dot(np.linalg.inv(grads), F_hat)
+
+ # update
+ self.us = all_us[::self.NUM_INPUT]
+ self.dummy_us = all_us[1::self.NUM_INPUT]
+ self.raws = all_us[2::self.NUM_INPUT]
+
+ # final insert
+ self.us = all_us[::self.NUM_INPUT]
+ self.dummy_us = all_us[1::self.NUM_INPUT]
+ self.raws = all_us[2::self.NUM_INPUT]
+
+ x_1s, x_2s, lam_1s, lam_2s = self.simulator.calc_predict_and_adjoint_state(x_1, x_2, self.us, self.N, dt)
+
+ F = self._calc_f(x_1s, x_2s, lam_1s, lam_2s, all_us, self.N, dt)
+
+ # print("check val of F = {0}".format(np.linalg.norm(F)))
+ # input()
+
+ # for save
+ self.history_f.append(np.linalg.norm(F))
+ self.history_u.append(self.us[0])
+ self.history_dummy_u.append(self.dummy_us[0])
+ self.history_raw.append(self.raws[0])
+
+ return self.us
+
+ def _calc_f(self, x_1s, x_2s, lam_1s, lam_2s, all_us, N, dt):
+ """
+ Parameters
+ ------------
+ x_1s : list of float
+ predicted x_1s for N steps
+ x_2s : list of float
+ predicted x_2s for N steps
+ lam_1s : list of float
+ adjoint state of x_1s, lam_1s for N steps
+ lam_2s : list of float
+ adjoint state of x_2s, lam_2s for N steps
+ us : list of float
+ estimated optimal system input
+ dummy_us : list of float
+ estimated dummy input
+ raws : list of float
+ estimated constraint variable
+ N : int
+ predict time step
+ dt : float
+ sampling time of system
+ """
+ F = []
+
+ us = all_us[::self.NUM_INPUT]
+ dummy_us = all_us[1::self.NUM_INPUT]
+ raws = all_us[2::self.NUM_INPUT]
+
+ for i in range(N):
+ F.append(0.5 * us[i] + lam_2s[i] + 2. * raws[i] * us[i])
+ F.append(-0.01 + 2. * raws[i] * dummy_us[i])
+ F.append(us[i]**2 + dummy_us[i]**2 - 0.5**2)
+
+ return np.array(F)
+
+def main():
+ # simulation time
+ dt = 0.01
+ iteration_time = 20.
+ iteration_num = int(iteration_time/dt)
+
+ # plant
+ plant_system = SampleSystem(init_x_1=2., init_x_2=0.)
+
+ # controller
+ controller = NMPCControllerWithNewton()
+
+ # for i in range(iteration_num)
+ for i in range(1, iteration_num):
+ print("iteration = {}".format(i))
+ time = float(i) * dt
+ x_1 = plant_system.x_1
+ x_2 = plant_system.x_2
+ # make input
+ us = controller.calc_input(x_1, x_2, time)
+ # update state
+ plant_system.update_state(us[0])
+
+ # figure
+ fig = plt.figure()
+
+ x_1_fig = fig.add_subplot(321)
+ x_2_fig = fig.add_subplot(322)
+ u_fig = fig.add_subplot(323)
+ dummy_fig = fig.add_subplot(324)
+ raw_fig = fig.add_subplot(325)
+ f_fig = fig.add_subplot(326)
+
+ x_1_fig.plot(np.arange(iteration_num)*dt, plant_system.history_x_1)
+ x_1_fig.set_xlabel("time [s]")
+ x_1_fig.set_ylabel("x_1")
+
+ x_2_fig.plot(np.arange(iteration_num)*dt, plant_system.history_x_2)
+ x_2_fig.set_xlabel("time [s]")
+ x_2_fig.set_ylabel("x_2")
+
+ u_fig.plot(np.arange(iteration_num - 1)*dt, controller.history_u)
+ u_fig.set_xlabel("time [s]")
+ u_fig.set_ylabel("u")
+
+ dummy_fig.plot(np.arange(iteration_num - 1)*dt, controller.history_dummy_u)
+ dummy_fig.set_xlabel("time [s]")
+ dummy_fig.set_ylabel("dummy u")
+
+ raw_fig.plot(np.arange(iteration_num - 1)*dt, controller.history_raw)
+ raw_fig.set_xlabel("time [s]")
+ raw_fig.set_ylabel("raw")
+
+ f_fig.plot(np.arange(iteration_num - 1)*dt, controller.history_f)
+ f_fig.set_xlabel("time [s]")
+ f_fig.set_ylabel("optimal error")
+
+ fig.tight_layout()
+
+ plt.show()
+
+
+if __name__ == "__main__":
+ main()
+
+
+